Quickstart#
import numpy as np
import bdms
Birth, death, and mutation processes#
We define a simple binary process.
birth = bdms.poisson.DiscreteProcess([1.0, 2.0])
death = bdms.poisson.ConstantProcess(1.0)
mutation = bdms.poisson.ConstantProcess(1.0)
mutator = bdms.mutators.DiscreteMutator((0, 1), np.array([[0, 1], [1, 0]]))
Simulation#
Initialize random number generator
rng = np.random.default_rng(seed=0)
Initialize a tree with a root node in state 0
tree = bdms.Tree(state=0)
Simulate for a specified time
time_to_sampling = 5.0
tree.evolve(
time_to_sampling,
birth_process=birth,
death_process=death,
mutation_process=mutation,
mutator=mutator,
seed=rng,
)
Randomly sample survivor tips
tree.sample_survivors(p=0.5, seed=rng)
Visualization#
Define keyword arguments that we’ll use repeatedly for tree visualization
viz_kwargs = dict(
color_map={0: "red", 1: "blue"},
h=5,
units="in",
)
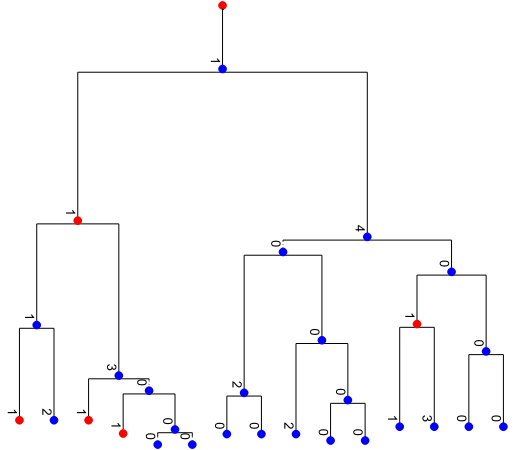
Render the full simulated tree
tree.render("%%inline", **viz_kwargs)
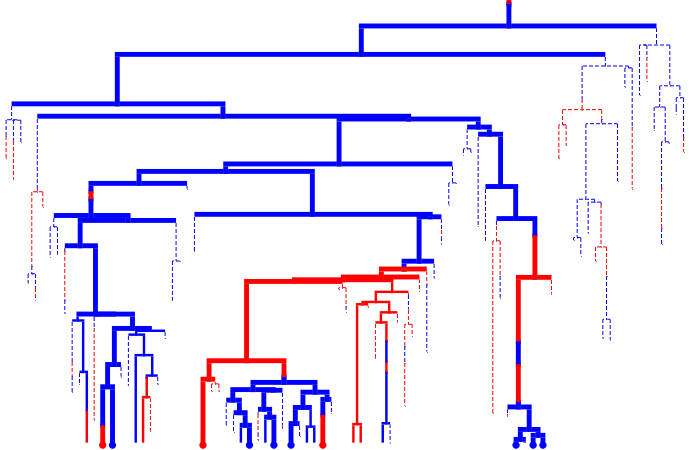
Partially observed trees#
Prune the tree to only include the ancestry of the sampled leaves
tree.prune_unsampled()
tree.render("%%inline", **viz_kwargs)
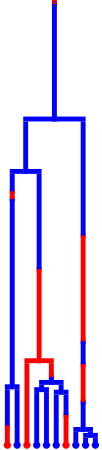
Also remove the mutation event unifurcations
tree.remove_mutation_events()
tree.render("%%inline", **viz_kwargs)
Inhomogeneous processes#
We subclass the abstract class bdms.poisson.InhomogeneousProcess.
To concretize, we must override its abstract method bdms.poisson.InhomogeneousProcess.λ_inhomogeneous().
We’ll use this for a death rate that linearly increases over time, and is independent of state.
class RampingProcess(bdms.poisson.InhomogeneousProcess):
def __init__(self, intercept, slope, *args, **kwargs):
self.intercept = intercept
self.slope = slope
super().__init__(*args, **kwargs)
def λ_inhomogeneous(self, x, t):
return self.intercept + self.slope * t
death = RampingProcess(1.0, 0.06)
Initialize tree as before
tree = bdms.Tree()
tree.state = 0
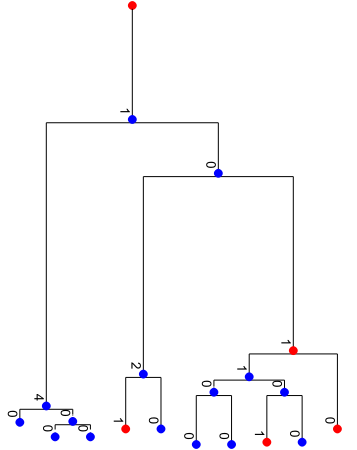
Simulate, sample, and visualize
tree.evolve(
time_to_sampling,
birth_process=birth,
death_process=death,
mutation_process=mutation,
mutator=mutator,
seed=rng,
)
tree.sample_survivors(p=0.5, seed=rng)
tree.render("%%inline", **viz_kwargs)
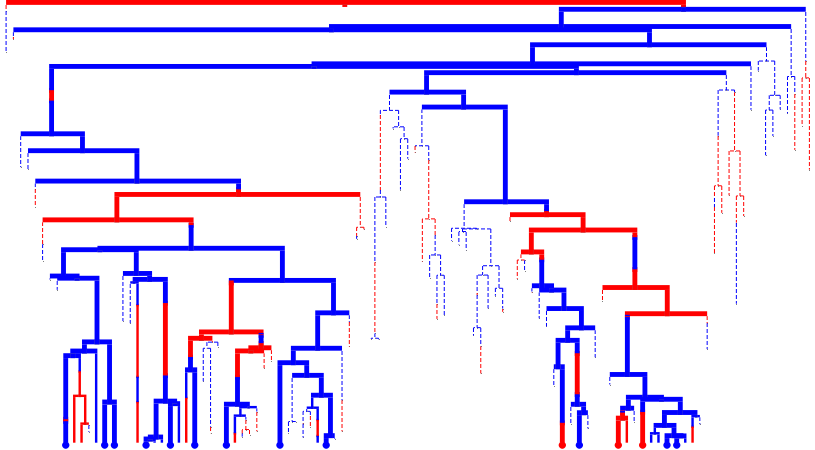
Prune and visualize
tree.prune_unsampled()
tree.render("%%inline", **viz_kwargs)
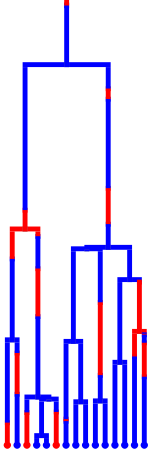
tree.remove_mutation_events()
tree.render("%%inline", **viz_kwargs)