Two preprints on inferring affinity-fitness response in germinal centers from trees
Two collaborative preprints investigate how fitness of affinity-maturing B cells depends on affinity changes in their mutating B-cell receptors, both using B-cell lineage trees from our recent GC (germinal center) replay preprint
-
Preprint led by Thanasi Bakis models trees via phylogenetic birth-death processes and performs Bayesian posterior inference of affinity-fitness response.
-
Preprint by Duncan Ralph uses the same trees but takes a simulation-based deep-learning approach to inferring parameters.
Update: Paper now out at eLife!
In the GC Replay paper we inferred an affinity-fitness landscape by modeling the time evolution of distributions of affinities (representing B-cells during GC evolution). Thanasi’s and Duncan’s papers both take a more fine-grained approach to this inference problem, leveraging the details of GC lineage trees with affinities assigned to nodes, rather than coarse-graining to affinity distributions.
“What good is this response function?”, you ask.
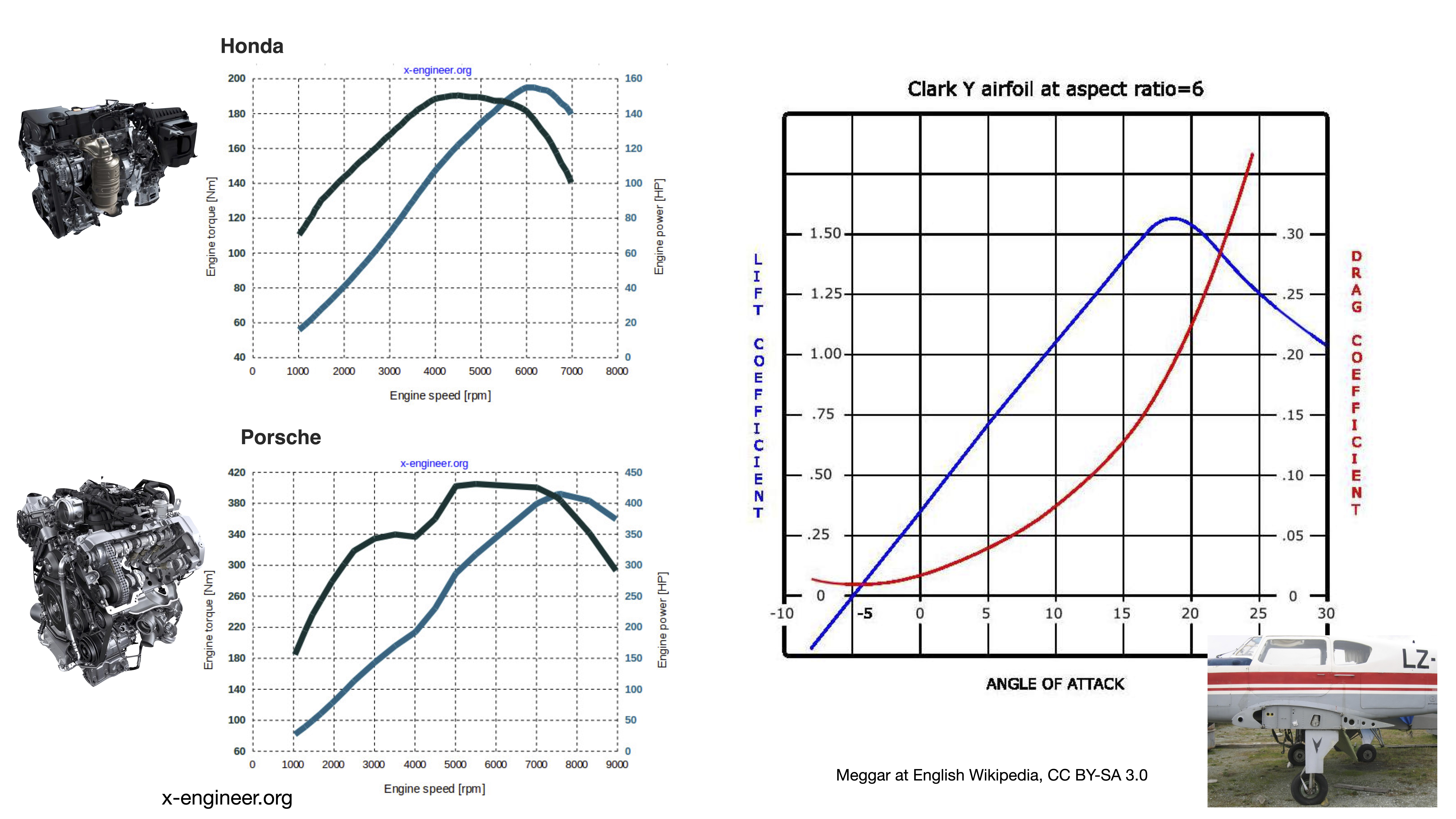
We have focused on inferring the shape of an affinity-fitness landscape, which may be dissatisfying if you want a mechanistic picture of how cells interact in micro detail to give rise to fitness. We think this coarse level of description is the appropriate goal, despite this lack of detail, and it’s a powerful tool. The affinity-fitness response function we infer is analogous to performance curves in engineering: a car engine can be described with a power band curve showing how torque and power vary with RPM, and an airplane can be described with airfoil curves describing how lift and drag vary with angle of attack. These give no insight about the mechanistic details of internal combustion engines, but they are predictive of performance: who wins a race, and who can fly. Likewise, the affinity-fitness response function is predictive of GC competitive dynamics: which mutations and lineages will win and lose, without specifying all micro-level details.

Comments